4.6. Power analysis by simulation#
We can think about theh Hypothesis Testing framework as a 2x2 grid - two possible states of reality and two (matching or non-matching) conclusions.
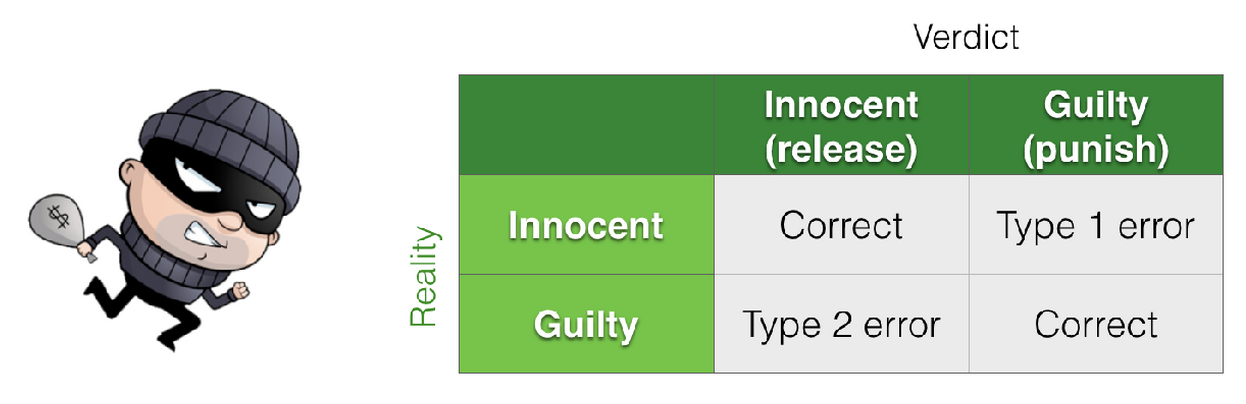
In general, null hypothesis testing concerns one state of reality - that is we assume the null is true
In contrast, power analysis concerns the other possible state of reality - we assume \(\mathcal{H_a}\) is true
In this notebook, focussing on an example of a correlation, we illustrate these hypothetical states of the world by generate two populations of ‘data’ from alternative realities:
A null population in which \(\mathcal{H_o}\) is true, and
A correlated population in which \(\mathcal{H_a}\) is true.
We will work with correlation (Pearson’s \(r\)), although later we will see that the same approach can be used for difference of means/ the \(t\)-test.
We can then:
work out the proportion of false positives we get if \(\mathcal{H_o}\) is true (by drawing many samples from the null population)
work out the proportion of false negatives we get if \(\mathcal{H_a}\) is true (by drawing many samples from the null population)
The latter gives us the power of the analysis.
Although we we later see that power analysis can be run using a built-in function in Python library statsmodels, it is useful to understand conceptually the simulation approach, as this gives an idea of what power analysis actually is
4.6.1. Video#
Here is a video summarizing the conceptual content of this page:
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/OnUrahxR_CI?si=6Hdw3NLGgYgOIfuz" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture; web-share" referrerpolicy="strict-origin-when-cross-origin" allowfullscreen></iframe>
Set up Python libraries#
As usual, run the code cell below to import the relevant Python libraries
# Set-up Python libraries - you need to run this but you don't need to change it
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats as stats
import pandas as pd
import seaborn as sns
sns.set_theme(style='white')
import statsmodels.api as sm
import statsmodels.formula.api as smf
import warnings
warnings.simplefilter('ignore', category=FutureWarning)
4.7. Example#
I collect data on end-of year exam scores in Maths and French for 50 high school studehts. Then I calculate the correlation coefficient, Pearson’s \(r\), between Maths and French scores across my sample of \(n=50\) participants.
\(\mathcal{H_0}\) Under the null hypothesis there is no correlation between maths scores and French scores
\(\mathcal{H_a}\) Under the alternative hypothesis, there is a positive correlation
This is a one-tailed test, at the \(\alpha=0.05\) level
I observe a correlation of \(r=0.25\) (this is my effect size)
Can I trust this result?
4.7.1. Eyeballing the data#
What does a correlation of 0.25 look like for 50 data points? Here are some examples of datasets that actually have r=0.25:
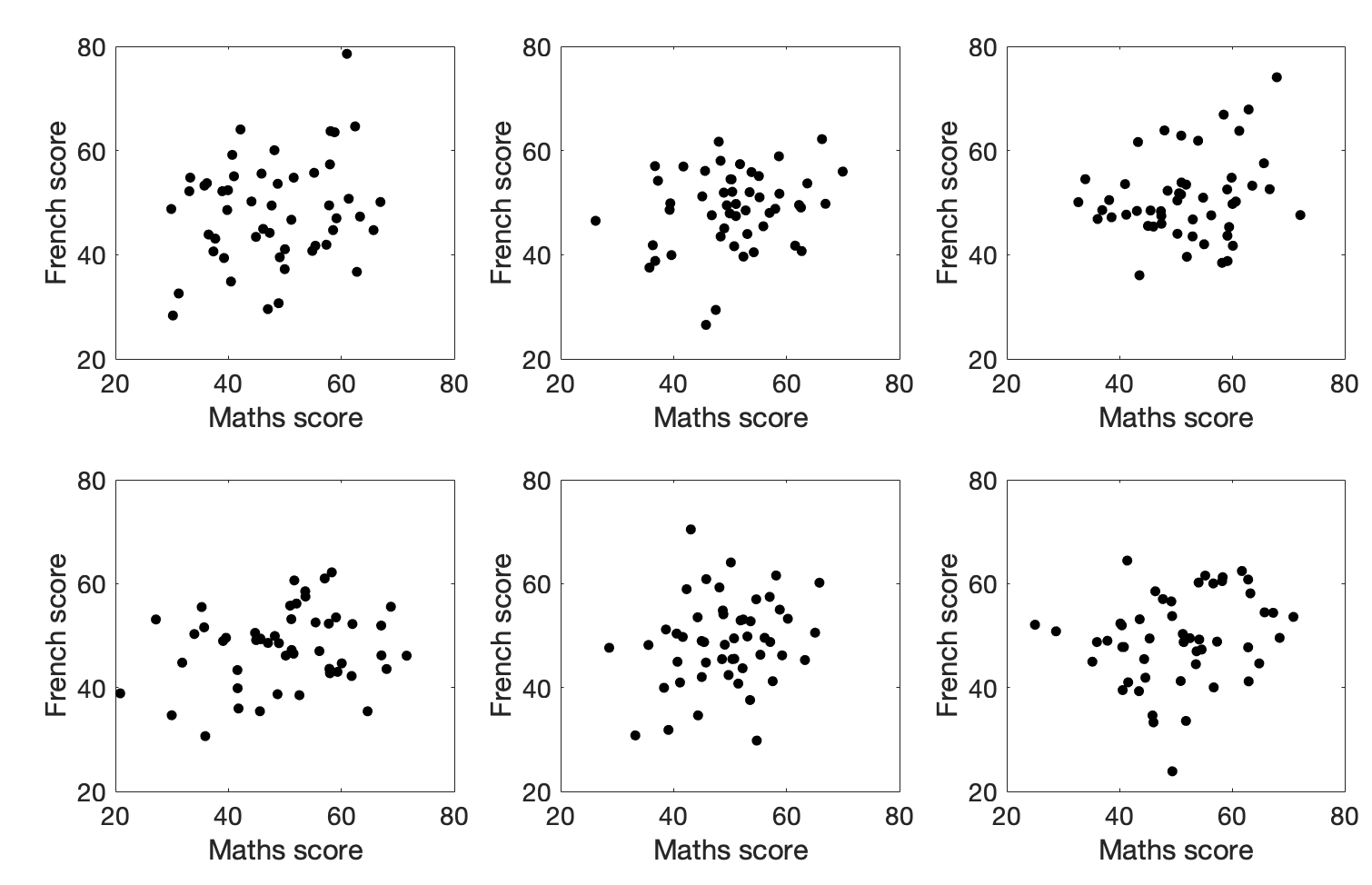
Would you trust those correlations?
4.7.2. Simulating the population#
A false positive (type I error) occurs when we get a significant correlation in a sample, even though there is no true correlation in the population
A false negative (type II error) occurs when we fail to get a significant correlation in a sample, even though there is a true correlation in the population
To understand the relationship between these two types of error, we are going to generate two ‘populations’ of data
A ‘null population’ where the true correlation \(\rho=0\)
A ‘correlated population’ where the true correlation \(\rho=0.25\)
NOTE - the symbol rho (\(\rho\)) denotes the true correlation in the population; it is the population equvalent of \(r\) in each sample. If \(\rho=0.25\), each sample will have \(r \approx 0.25\) but \(r\) will be slightly different in each sample.
Each dataset will have 25000 pairs in it (so 2 columns, 25000 rows), with a correlation between columns of zero (null population) or 0.25 (correlated population).
By drawing random samples of size \(n=50\) from each, we can ask:
how often do we find a significant correlation when drawing from the null population (false positive, Type I error)?
how often do we fail to find a significant correlation when drawing from the correlated population (false negative, Type II error)?
You will not be asked to replicate this code so don’t worry about the Python details
4.7.3. Null population#
# Code to generate the population with zero correlation - don't worry about understanding this bit
rho=0 # the desired correlation
# desired mean and sd of variables in columns 0 and 1
m0=50
m1=50
s0=8
s1=5
c=[[1,rho],[rho,1]] # the correlation matrix that we want in the simulated dataset
# make two columns of numbers with the desired correlation (mean of each column is zero, sd is 1)
tmp=np.random.multivariate_normal([0,0],c,size=25000)
# scale the variables to get correct mean and sd
tmp[:,0]=(tmp[:,0]*s0)+m0
tmp[:,1]=(tmp[:,1]*s1)+m1
pop_rZero=pd.DataFrame(tmp,columns=['Maths','French'])
# plot scatterplot
sns.scatterplot(data=pop_rZero,x='Maths',y='French',alpha=0.01)
plt.show()
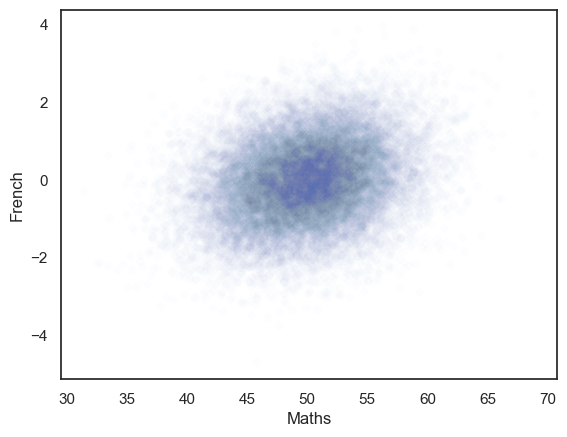
The generated dataset has two columns and 25,000 rows.
The correlation between the two columns should be close to the r value we set, ie zero.
The scatterplot definitely looks like there is no correlation!
# check that the actual correlation is in the 10000 simulated samples
pop_rZero.corr()
| Maths | French | |
|---|---|---|
| Maths | 1.000000 | 0.005595 |
| French | 0.005595 | 1.000000 |
Simulating false positives#
We can now work out how often we get a significant correlation (\(p<0.05\)) in samples (\(n=50\)) drawn from the null population, by drawing 10000 samples and getting the p value for each one using stats.pearsonr().pvalue.
The number of false positives will simply be the proportion of those obtained \(p\)-values that are below 5%.
nReps=10000
sampleSize=50
p = np.empty(nReps)
for i in range(nReps):
sample = pop_rZero.sample(n=sampleSize) # sample 50 random rows from our null population
p[i] = stats.pearsonr(sample['Maths'], sample['French'], alternative='greater').pvalue # correlate and get p-value
# How many of our 10000 samples had p<0.05?
np.mean(p<0.05)
0.1046
Hopefully the proportion of samples from the no-correlation distribution with p<0.05 is about…. 0.05.
4.7.5. Determining the required sample size#
We have just seen that a large sample size is needed to get good power for a weak correlation. How large, exactly?
We can loop over different sample sizes and plot how power increases as sample size increases:
Warning - if you run this code block it may take a couple of minutes, or longer if your computer is a bit slow. You don’t actually need to run it.
n=range(2,150,10)
power = np.empty(len(n))
for s in range(len(n)):
nReps=1000
p = np.empty(nReps)
sampleSize=n[s]
for i in range(nReps):
sample = pop_rNonZero.sample(n=sampleSize)
p[i] = stats.pearsonr(sample['Maths'], sample['French'], alternative='greater').pvalue
power[s]=np.mean(p<0.05)
# plot the results
plt.plot(n,power,'k.-')
plt.plot([min(n),max(n)],[0.8,0.8],'r--') # reference line for 80% power
plt.xlabel('sample size')
plt.ylabel('power')
plt.show()
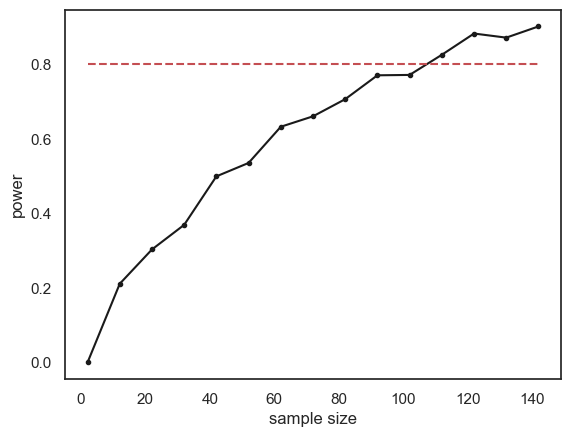
The relationship between sample size and power is slightly bumpy as we checked only 1000 simulated samples at each sample size. If you increased nReps to 10,000 the curve would look smoother (but the simulation would take a long time to run).
Conventionally a power of 80% (indicated by the red dashed line on the graph) is considered desirable (80% is a completely arbitrary value by the way).
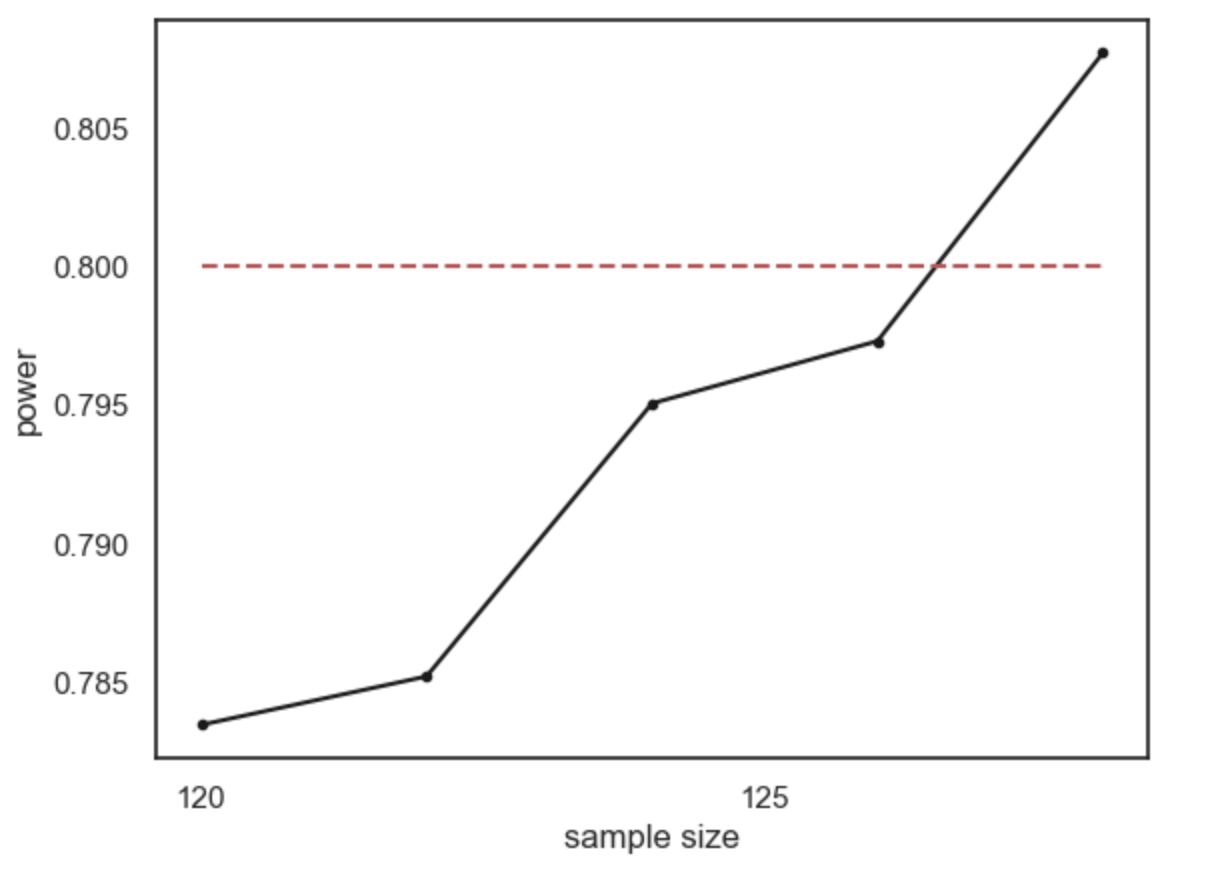
What sample size do we need to ge t power of 80%? It looks from the graph like you would need around n=120. Here are the results if we run a finer grained simulation in that range:
4.7.6. Recap#
Power is defined as the propotion of the time a significant effect is detected, given that there is an effect (of a certain size in the data)
Power depends on sample size; a larger sample gives a better chance to detect the effect (if there is one)
To reach a desirable level of power (say 80%) we need a certain sample size; this sample size depends on the effect size and \(\alpha\) value we are working with
The smaller the effect size, the larger the sample size neeed to detect it.
The more stringent the \(\alpha\) value, the larger the sample needed to detect an effect of a given size
To work out the required sample size, we did the following steps for different values of \(n\):
simulated data with the effect size we think is present in the population (\(\rho=0.25\))
drew many samples of size \(n\) and tested for a significant effect
worked out the power as the proportion of samples with a significant effect
The process of determining the required sample size to detect a certain effect with a certain power is called power analysis
4.7.7. Built in function for power#
Now you understand conceptually what power analysis is about, we can move on to running it using a built-in function.
There is a built in function in a package called statsmodels for doing power analysis.
In the next notebook we will see this for \(t\)-tests and correlations.